
Tight turns and loose, flexible loops link the more "regular" secondary structure elements. Other extended structures such as the polyproline helix and alpha sheet are rare in native state proteins but are often hypothesized as important protein folding intermediates. Other helices, such as the 3 10 helix and π helix, are calculated to have energetically favorable hydrogen-bonding patterns but are rarely observed in natural proteins except at the ends of α helices due to unfavorable backbone packing in the center of the helix.

The most common secondary structures are alpha helices and beta sheets.
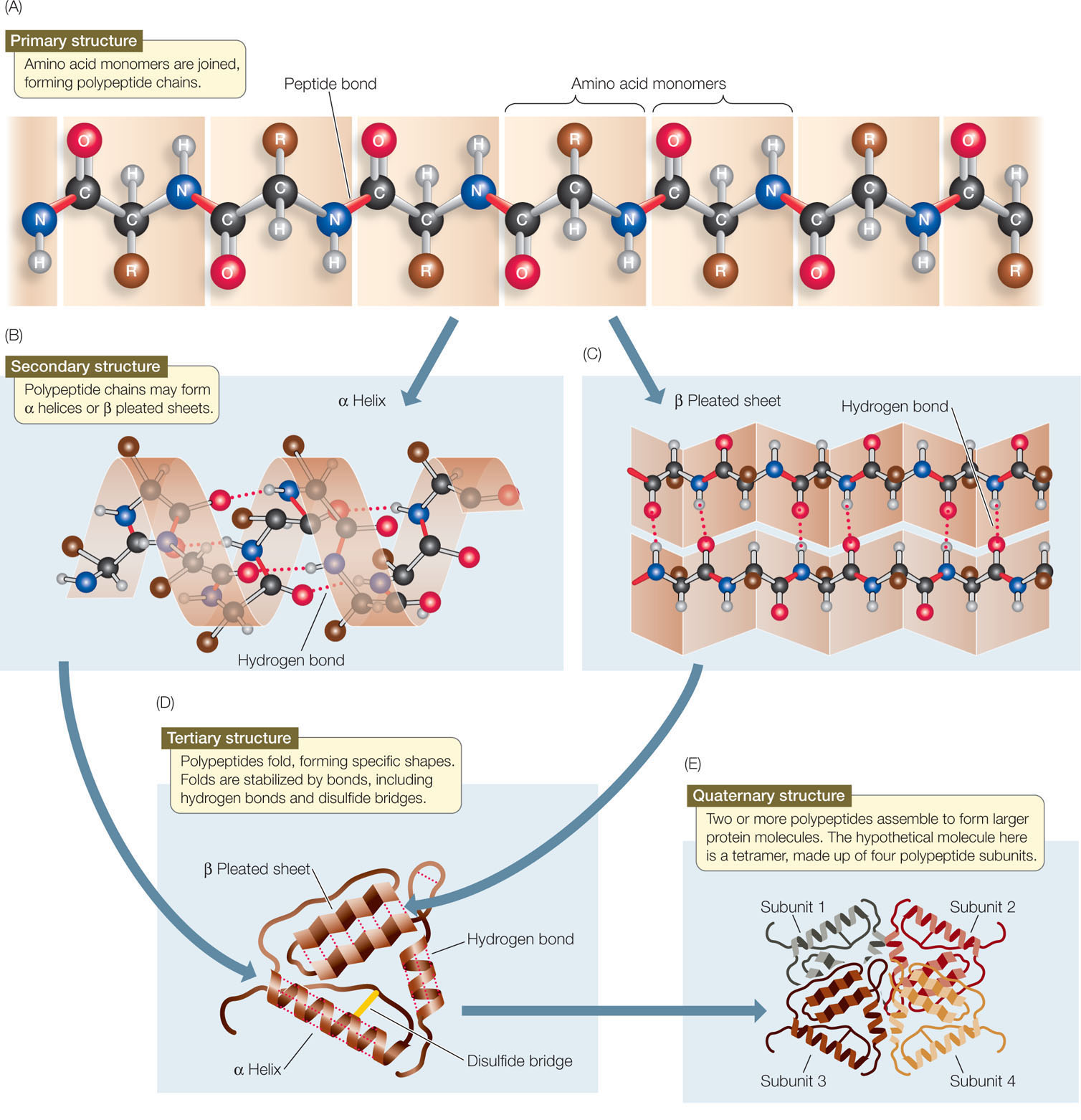
Cartoon above, atoms below with nitrogen in blue, oxygen in red ( PDB: 1AXC) Interactive diagram of hydrogen bonds in protein secondary structure. Types Structural features of the three major forms of protein helices Geometry attribute Other types of biopolymers such as nucleic acids also possess characteristic secondary structures. The concept of secondary structure was first introduced by Kaj Ulrik Linderstrøm-Lang at Stanford in 1952. Secondary structure may alternatively be defined based on the regular pattern of backbone dihedral angles in a particular region of the Ramachandran plot regardless of whether it has the correct hydrogen bonds. Secondary structure is formally defined by the pattern of hydrogen bonds between the amino hydrogen and carboxyl oxygen atoms in the peptide backbone. Secondary structure elements typically spontaneously form as an intermediate before the protein folds into its three dimensional tertiary structure. The two most common secondary structural elements are alpha helices and beta sheets, though beta turns and omega loops occur as well.
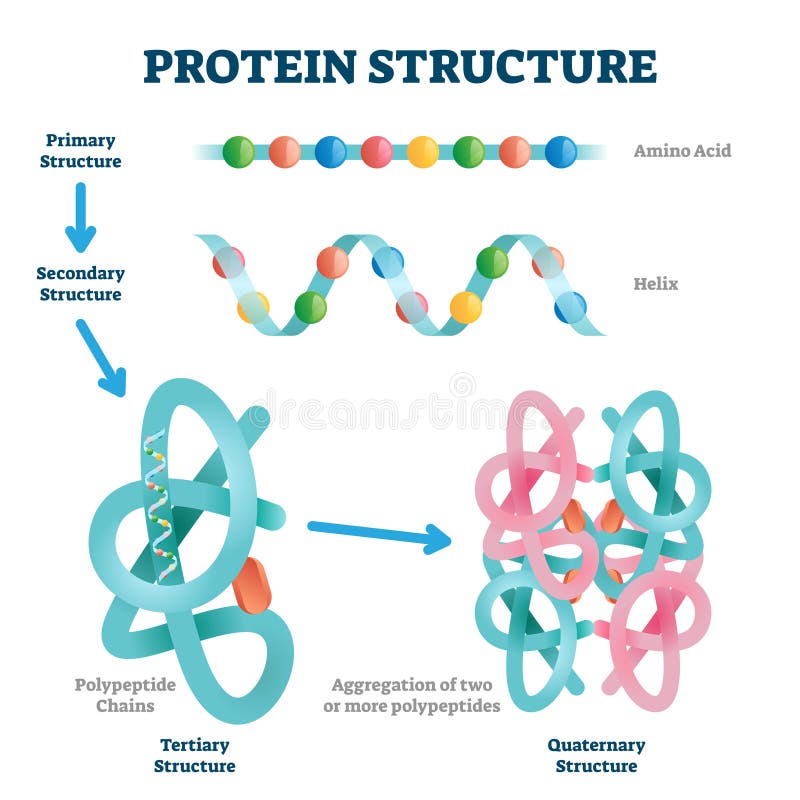
Protein secondary structure is the local spatial conformation of the polypeptide backbone excluding the side chains. This diagram (which is interactive) of protein structure uses PCNA as an example.


 0 kommentar(er)
0 kommentar(er)
